This is The Most Complete Guide to PySpark DataFrame Operations. A bookmarkable cheatsheet containing all the Dataframe Functionality you might need. In this post we will talk about installing Spark, standard Spark functionalities you will need to work with DataFrames, and finally some tips to handle the inevitable errors you will face.
Installation
We will be working on installing Spark on Ubuntu 18.04, but the steps should remain the same for MAC too. Assuming that you already have Anaconda and Python3 installed. After that, you can just go through these steps:
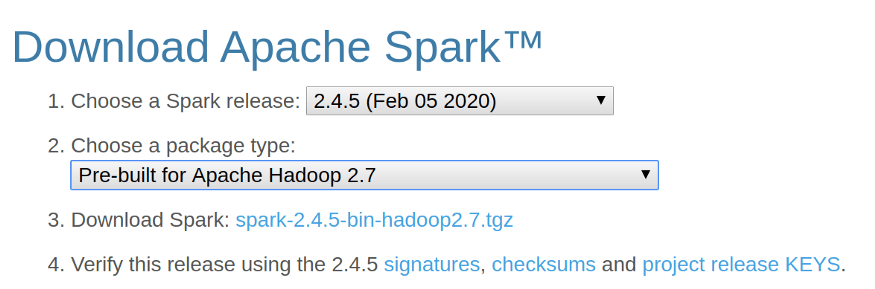
- Download the Spark Binary from the Apache Spark Website. And click on the Download Spark link to download Spark.
- Once you have downloaded the above file, you can start with unzipping the file in your home directory. Just Open up the terminal and put these commands in.
cd ~
cp Downloads/spark-2.4.5-bin-hadoop2.7.tgz ~
tar -zxvf spark-2.4.5-bin-hadoop2.7.tgz- Check your Java Version. As of version 2.4 Spark works with Java 8. You can check your Java Version using the command
java -versionon the terminal window.
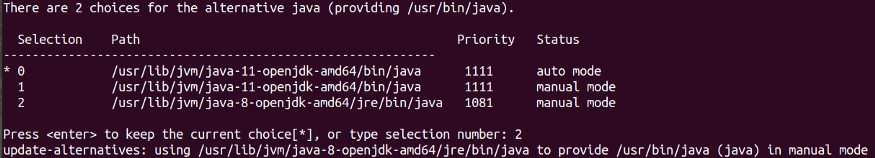
I had Java 11 in my machine, so I had to run the following commands on my terminal to install and change default Java to Java 8:
sudo apt install openjdk-8-jdk
sudo update-alternatives --config javaYou will need to manually select Java version 8 by typing the selection number.
- Edit your
~/.bashrcfile and add the following lines at the end of the file. This is just to add Spark directory into $PATH:
function pysparknb ()
{
#Spark path
SPARK_PATH=~/spark-2.4.5-bin-hadoop2.7
export PYSPARK_DRIVER_PYTHON="jupyter"
export PYSPARK_DRIVER_PYTHON_OPTS="notebook"
# For pyarrow 0.15 users, you have to add the line below or you will get an error while using pandas_udf
export ARROW_PRE_0_15_IPC_FORMAT=1# Change the local[10] to local[numCores in your machine]
$SPARK_PATH/bin/pyspark --master local[10]
}- Source
~/.bashrc
source ~/.bashrc- Run the
pysparknbfunction in the terminal and you will be able to access the notebook. You will be able to open a new notebook as well as thesparkcontextwill be loaded automatically.
pysparknbData
With the installation out of the way, we can move to the more interesting part of this post. I will be working with the Data Science for COVID-19 in South Korea, which is one of the most detailed datasets on the internet for COVID.
Please note that I will be using this dataset to showcase some of the most useful functionalities of Spark, but this should not be in any way considered a data exploration exercise for this amazing dataset.
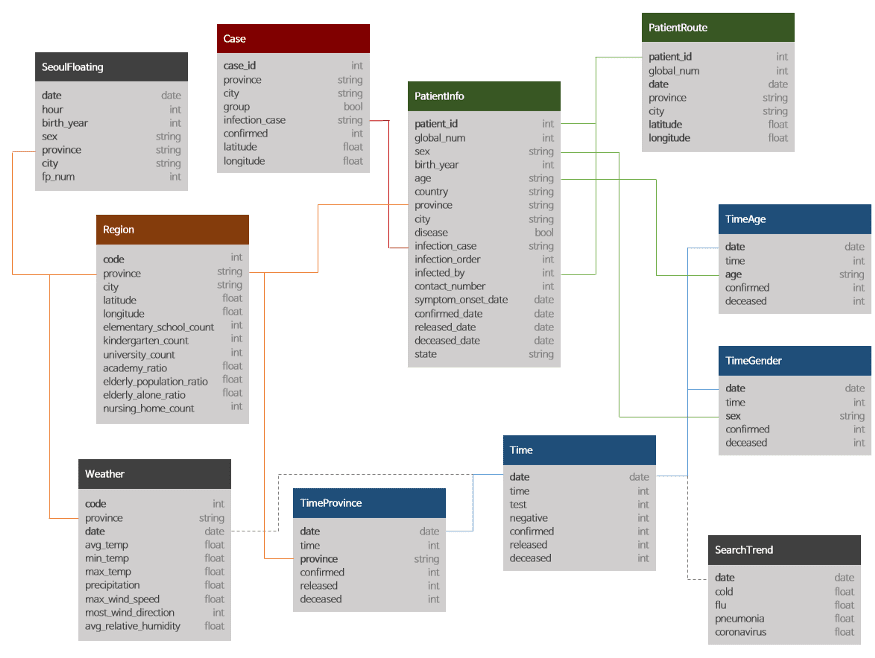
I will mainly work with the following three tables only in this post:
- Cases
- Region
- TimeProvince
1. Basic Functions
Read
We can start by loading the files in our dataset using the spark.read.load command. This command reads parquet files, which is the default file format for spark, but you can add the parameter format to read .csv files using it.
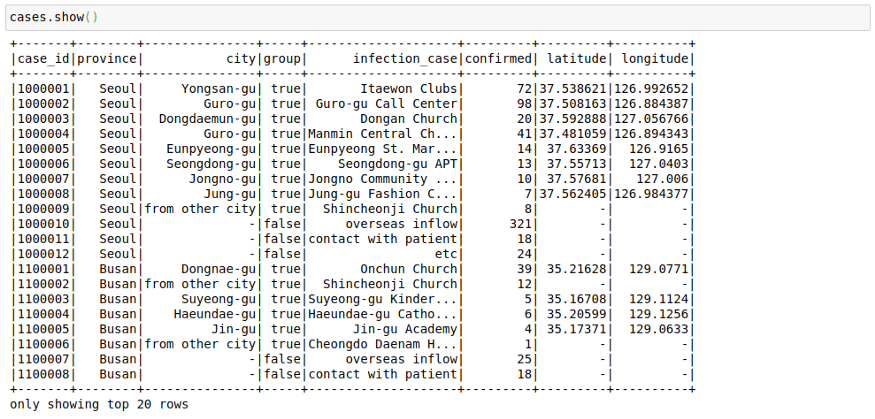
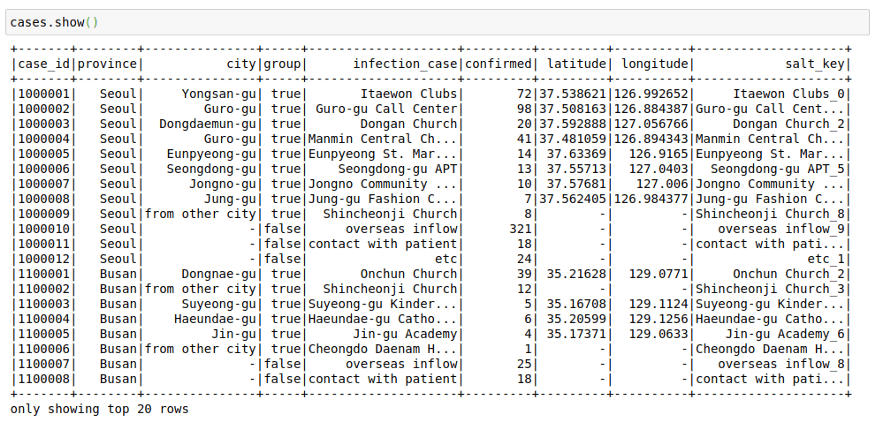
cases = spark.read.load("/home/mytechmint/projects/sparkdf/coronavirusdataset/Case.csv",format="csv", sep=",", inferSchema="true", header="true")See a few rows in the file
cases.show()This file contains the cases grouped by way of the infection spread. This might have helped in the rigorous tracking of Corona Cases in South Korea.
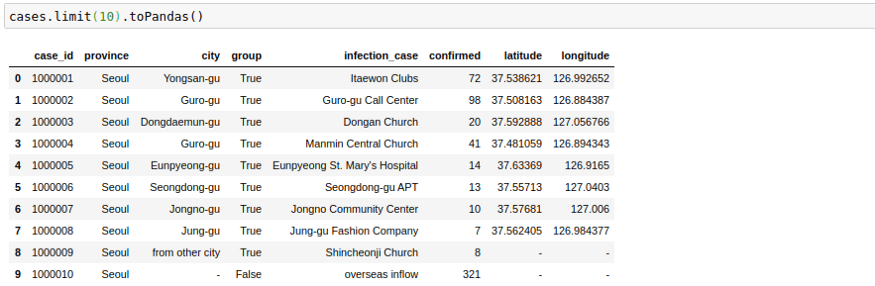
The way this file looks is great right now, but sometimes as we increase the number of columns, the formatting becomes not too great. I have noticed that the following trick helps in displaying in pandas format in my Jupyter Notebook. The .toPandas() the function converts a spark data frame into a pandas Dataframe which is easier to show.
cases.limit(10).toPandas()Change Column Names
Sometimes we would like to change the name of columns in our Spark Dataframes. We can do this simply using the below command to change a single column:
cases = cases.withColumnRenamed("infection_case","infection_source")Or for all columns:
cases = cases.toDF(*['case_id', 'province', 'city', 'group', 'infection_case', 'confirmed',
'latitude', 'longitude'])Select Columns
We can select a subset of columns using the select keyword.
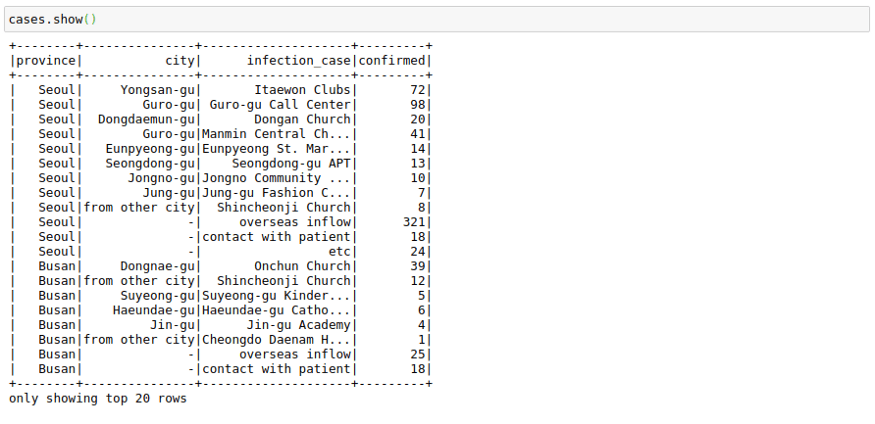
cases = cases.select('province','city','infection_case','confirmed')
cases.show()Sort
We can sort by the number of confirmed cases. Here note that the cases data frame will not change after performing this command as we don’t assign it to any variable.
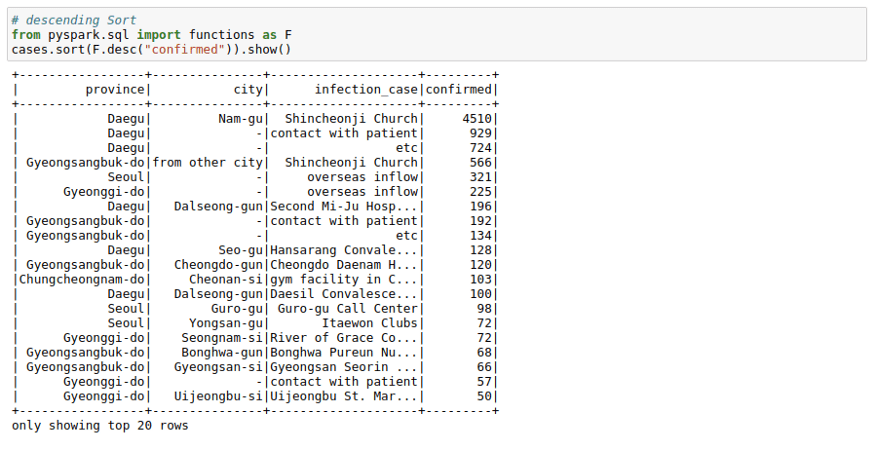
cases.sort("confirmed").show()But that is inverted. We want to see most cases at the top. We can do this using the F.desc function:
# Descending Sort
from pyspark.sql import functions as F
cases.sort(F.desc("confirmed")).show()We can see that most cases in a logical area in South Korea originated from `Shincheonji Church`.
Cast
Though we don’t face it in this dataset, there might be scenarios where Pyspark reads a double as integer or string, In such cases, you can use the cast function to convert types.
from pyspark.sql.types import DoubleType, IntegerType, StringType
cases = cases.withColumn('confirmed', F.col('confirmed').cast(IntegerType()))
cases = cases.withColumn('city', F.col('city').cast(StringType()))Filter
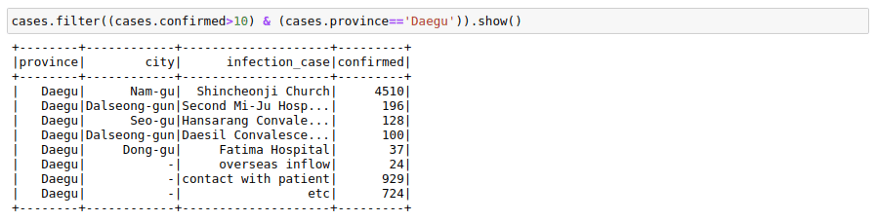
We can filter a data frame using multiple conditions using AND(&), OR(|) and NOT(~) conditions. For example, we may want to find out all the different infection_case in Daegu Province with more than 10 confirmed cases.
cases.filter((cases.confirmed>10) & (cases.province=='Daegu')).show()GroupBy
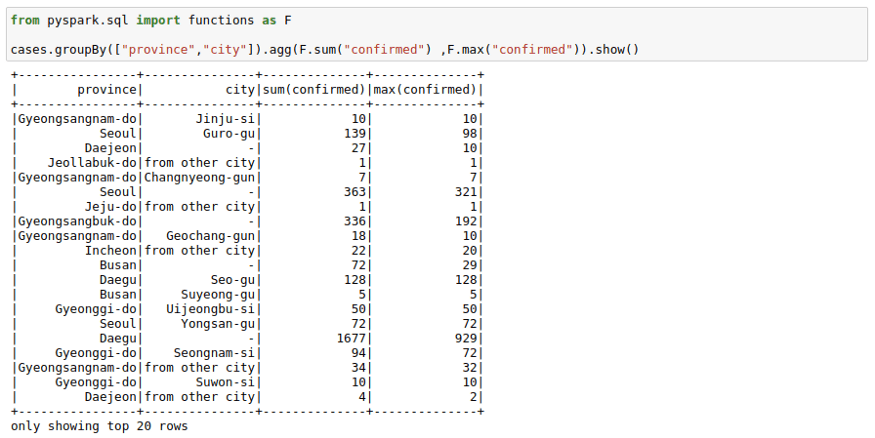
We can use groupBy function with a spark DataFrame too. Pretty much same as the pandas groupBy with the exception that you will need to import pyspark.sql.functions. Here is the list of functions you can use with this function module.
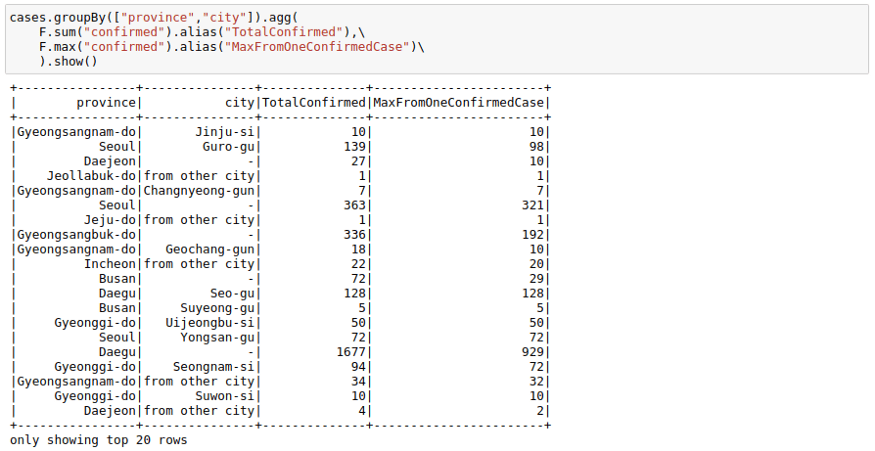
cases.groupBy(["province","city"]).agg(F.sum("confirmed") ,F.max("confirmed")).show()If you don’t like the new column names, you can use the alias keyword to rename columns in the agg command itself.
cases.groupBy(["province","city"]).agg(
F.sum("confirmed").alias("TotalConfirmed"),\
F.max("confirmed").alias("MaxFromOneConfirmedCase")\
).show()Joins
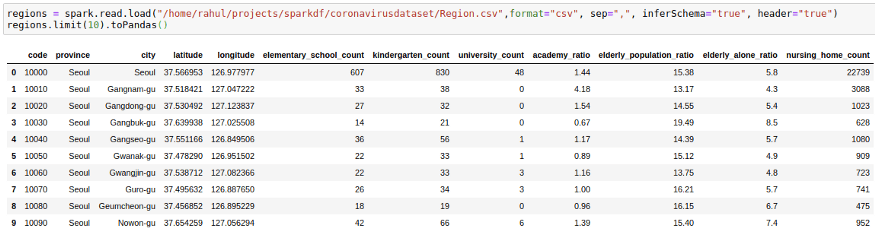
To Start with Joins we will need to introduce one more CSV file. We will go with the region file which contains region information such as elementary_school_count, elderly_population_ratio, etc.
regions = spark.read.load("/home/mytechmint/projects/sparkdf/coronavirusdataset/Region.csv",format="csv", sep=",", inferSchema="true", header="true")
regions.limit(10).toPandas()We want to get this information in our cases file by joining the two DataFrames. We can do this by using:
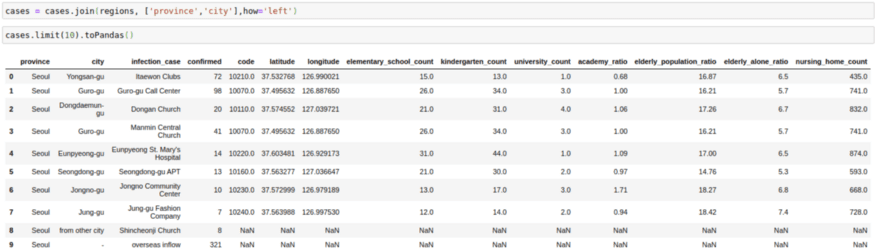
cases = cases.join(regions, ['province','city'],how='left')
cases.limit(10).toPandas()2. Broadcast/Map Side Joins
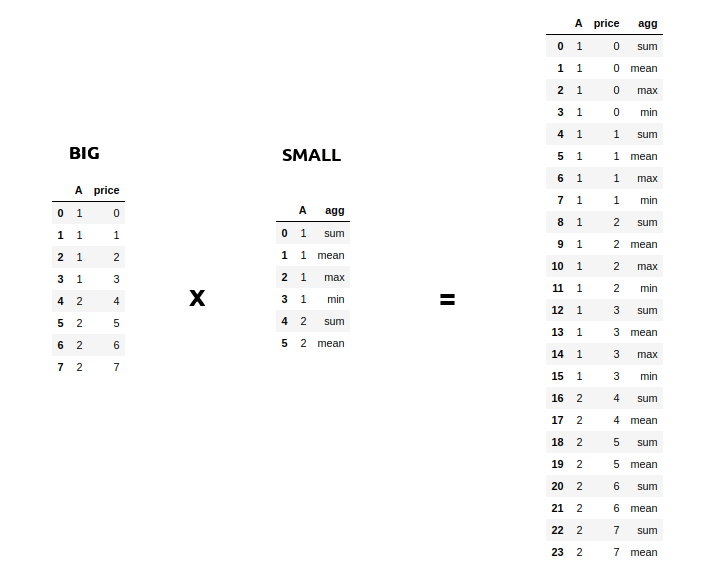
Sometimes you might face a scenario where you need to join a very big table(~1B Rows) with a very small table(~100–200 rows). The scenario might also involve increasing the size of your database as in the example below.
Such sort of operations is aplenty in Spark where you might want to apply multiple operations to a particular key. But assuming that the data for each key in the Big table is large, it will involve a lot of data movement. And sometimes so much that the application itself breaks. A small optimization then you can do when joining on such big tables(assuming the other table is small) is to broadcast the small table to each machine/node when you perform a join. You can do this easily using the broadcast keyword. This has been a lifesaver many times with Spark when everything else fails.
from pyspark.sql.functions import broadcast
cases = cases.join(broadcast(regions), ['province','city'],how='left')3. Use SQL with DataFrames
If you want, you can also use SQL with data frames. Let us try to run some SQL on the cases table.
We first register the cases data frame to a temporary table cases_table on which we can run SQL operations. As you can see, the result of the SQL select statement is again a Spark Dataframe.
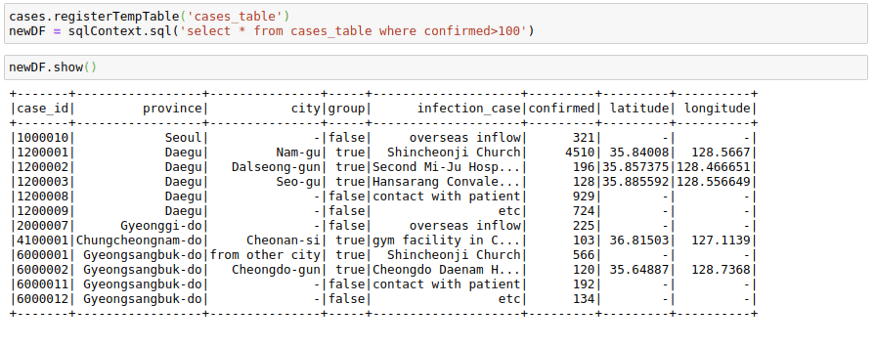
cases.registerTempTable('cases_table')
newDF = sqlContext.sql('select * from cases_table where confirmed>100')
newDF.show()I have shown a minimal example above, but you can use pretty much complex SQL queries involving GROUP BY, HAVING, AND ORDER BY clauses as well as aliases in the above query.
4. Create New Columns
There are many ways that you can use to create a column in a PySpark Dataframe. I will try to show the most usable of them.
Using Spark Native Functions
The best way to create a new column in a PySpark DataFrame is by using built-in functions. This is the most performant programmatical way to create a new column, so this is the first place I go whenever I want to do some column manipulation.
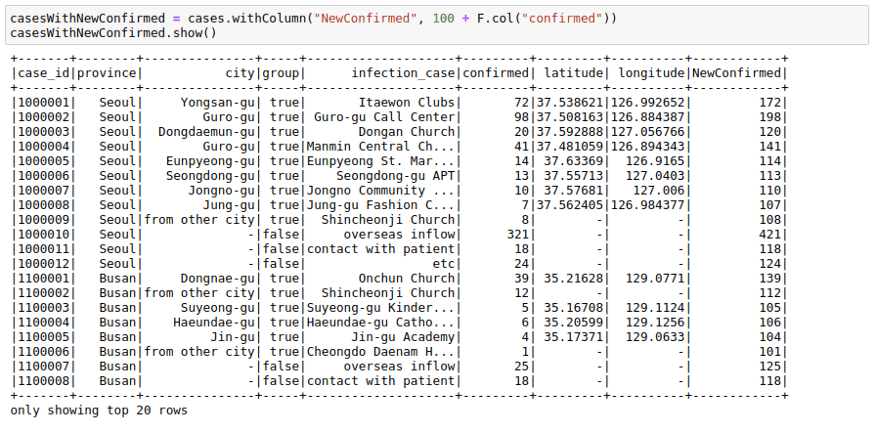
We can use .withcolumn along with PySpark SQL functions to create a new column. In essence, you can find String functions, Date functions, and Math functions already implemented using Spark functions. Our first function, the F.col function gives us access to the column. So if we wanted to add 100 to a column, we could use F.col as:
import pyspark.sql.functions as F
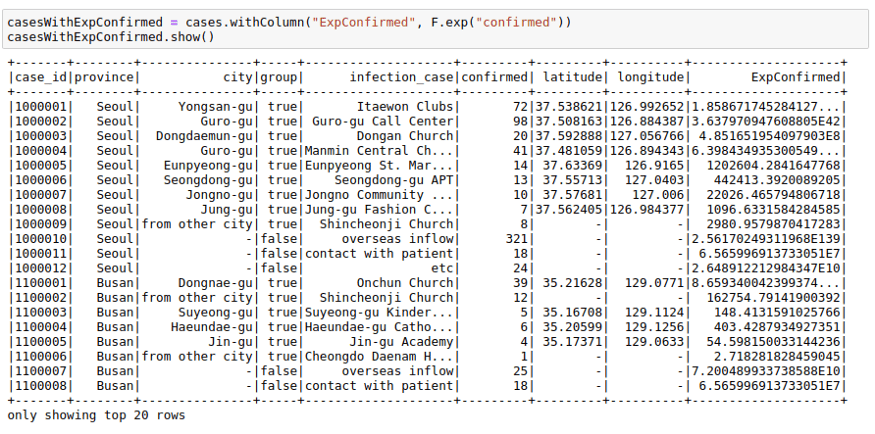
casesWithNewConfirmed = cases.withColumn("NewConfirmed", 100 + F.col("confirmed"))casesWithNewConfirmed.show()We can also use math functions like F.exp function:
casesWithExpConfirmed.show()To deep dive into more details of PySpark, please check PySpark Tutorial section.
There are a lot of other functions provided in this module, which are enough for most simple use cases. You can check out the functions list here.
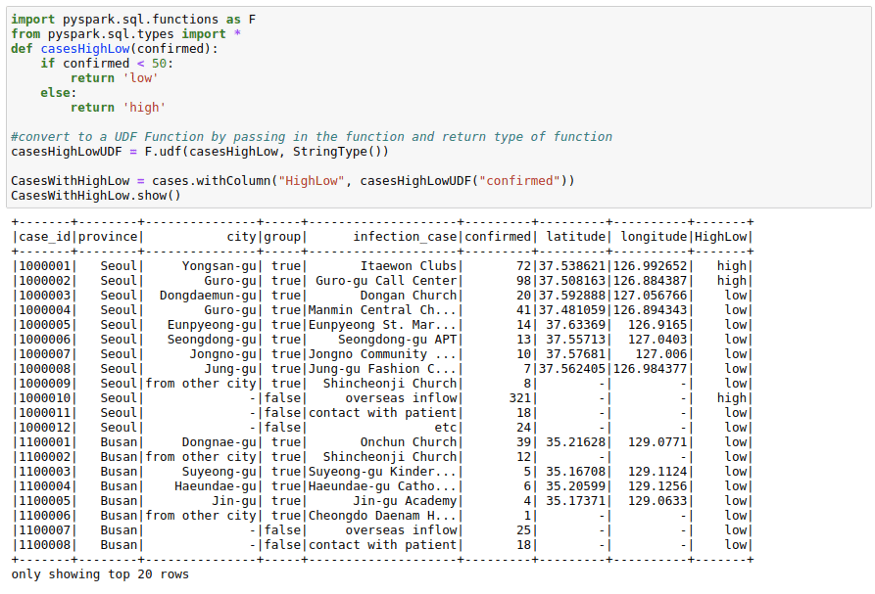
Using Spark UDFs
Sometimes we want to do complicated things to a column or multiple columns. This could be thought of as a map operation on a PySpark Dataframe to a single column or multiple columns. While Spark SQL functions do solve many use cases when it comes to column creation, I use Spark UDF whenever I need more matured Python functionality.
To use Spark UDFs, we need to use the F.udf function to convert a regular python function to a Spark UDF. We also need to specify the return type of the function. In this example the return type is StringType()
import pyspark.sql.functions as F
from pyspark.sql.types import *
def casesHighLow(confirmed):
if confirmed < 50:
return 'low'
else:
return 'high'
#convert to a UDF Function by passing in the function and return type of function
casesHighLowUDF = F.udf(casesHighLow, StringType())CasesWithHighLow = cases.withColumn("HighLow", casesHighLowUDF("confirmed"))
CasesWithHighLow.show()Using RDDs
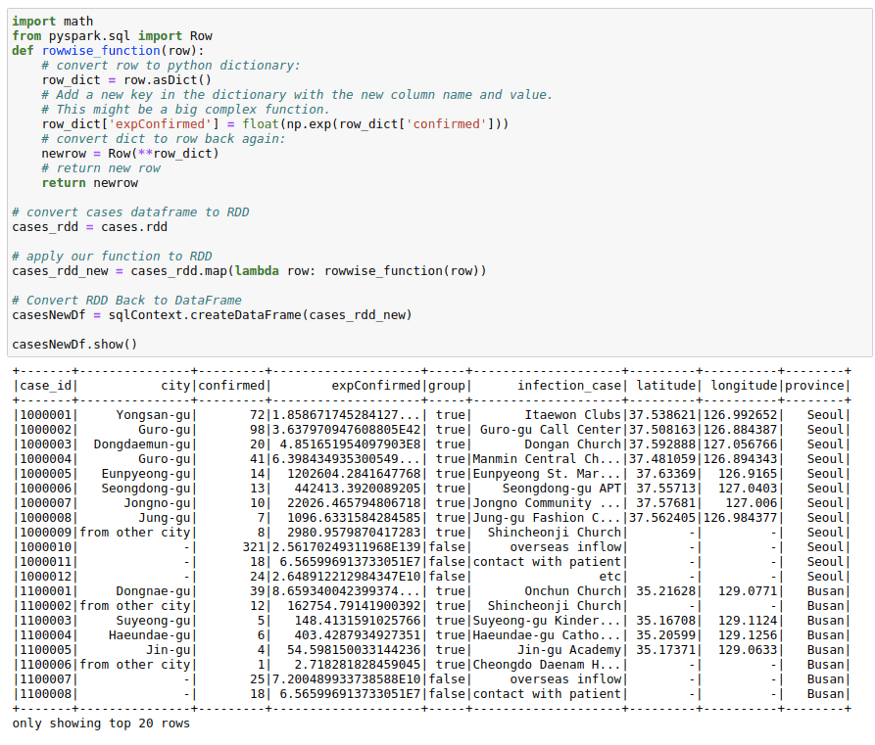
This might seem a little odd, but sometimes both the spark UDFs and SQL functions are not enough for a particular use-case. I have observed the RDDs being much more performant in some use-cases in real life. You might want to utilize the better partitioning that you get with spark RDDs. Or you may want to use group functions in Spark RDDs.
Whatever the case be, I find this way of using RDD to create new columns pretty useful for people who have experience working with RDDs that is the basic building block in the Spark ecosystem. Don’t worry much if you don’t understand it. It is just here for completion.
The process below makes use of the functionality to convert between Row and pythondict objects. We convert a row object to a dictionary. Work with the dictionary as we are used to and convert that dictionary back to row again. This might come in handy in a lot of situations.
import math
from pyspark.sql import Rowdef rowwise_function(row):
# convert row to python dictionary:
row_dict = row.asDict()
# Add a new key in the dictionary with the new column name and value.
# This might be a big complex function.
row_dict['expConfirmed'] = float(np.exp(row_dict['confirmed']))
# convert dict to row back again:
newrow = Row(**row_dict)
# return new row
return newrow# convert cases dataframe to RDD
cases_rdd = cases.rdd# apply our function to RDD
cases_rdd_new = cases_rdd.map(lambda row: rowwise_function(row))# Convert RDD Back to DataFrame
casesNewDf = sqlContext.createDataFrame(cases_rdd_new)casesNewDf.show()
Using Pandas UDF
This functionality was introduced in Spark version 2.3.1. And this allows you to use pandas functionality with Spark. I generally use it when I have to run a groupBy operation on a Spark data frame or whenever I need to create rolling features and want to use Pandas rolling functions/window functions rather than Spark window functions which we will go through later in this post.
The way we use it is by using the F.pandas_udf decorator. We assume here that the input to the function will be a pandas data frame. And we need to return a pandas data frame in turn from this function.
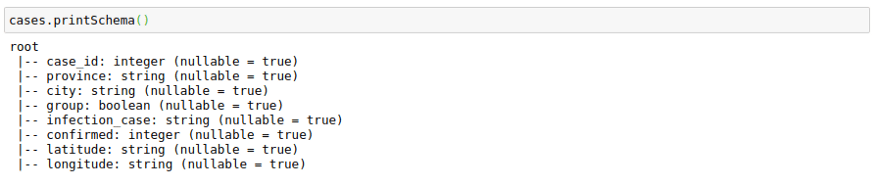
The only complexity here is that we have to provide a schema for the output Dataframe. We can use the original schema of a data frame to create the outSchema.
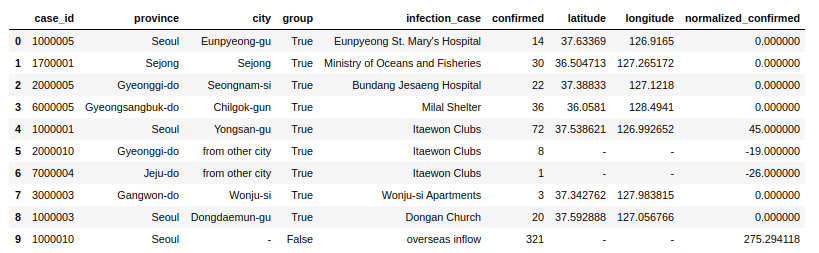
cases.printSchema()Here I am using Pandas UDF to get normalized confirmed cases grouped by infection_case. The main advantage here is that I get to work with pandas data frames in Spark.
from pyspark.sql.types import IntegerType, StringType, DoubleType, BooleanType
from pyspark.sql.types import StructType, StructField
# Declare the schema for the output of our function
outSchema = StructType([StructField('case_id',IntegerType(),True),
StructField('province',StringType(),True),
StructField('city',StringType(),True),
StructField('group',BooleanType(),True),
StructField('infection_case',StringType(),True),
StructField('confirmed',IntegerType(),True),
StructField('latitude',StringType(),True),
StructField('longitude',StringType(),True),
StructField('normalized_confirmed',DoubleType(),True)
])
# decorate our function with pandas_udf decorator
@F.pandas_udf(outSchema, F.PandasUDFType.GROUPED_MAP)
def subtract_mean(pdf):
# pdf is a pandas.DataFrame
v = pdf.confirmed
v = v - v.mean()
pdf['normalized_confirmed'] = v
return pdf
confirmed_groupwise_normalization = cases.groupby("infection_case").apply(subtract_mean)
confirmed_groupwise_normalization.limit(10).toPandas()To deep dive into more details of Pandas, please check the Pandas Tutorial section.
5. Spark Window Functions
Window functions may make a whole blog post in itself. Here I will talk about some of the most important window functions available in spark.
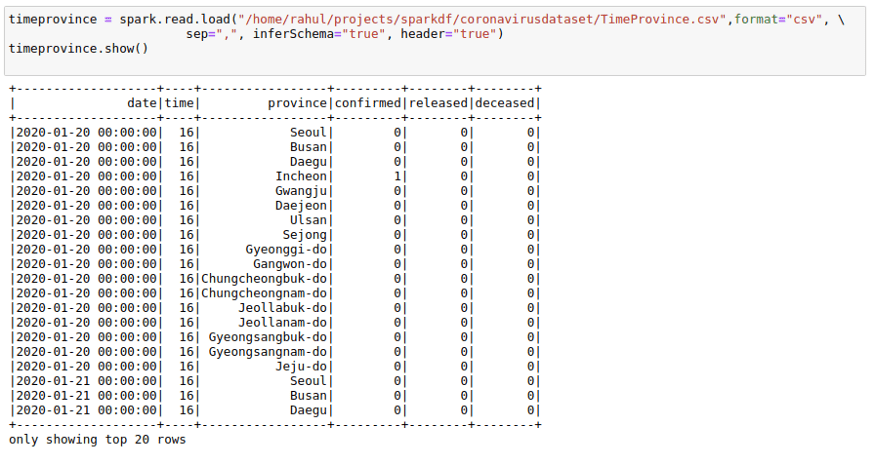
For this, I will also use one more data CSV, which has dates present as that will help with understanding Window functions much better. I will use the TimeProvince data frame which contains daily case information for each province.
Ranking
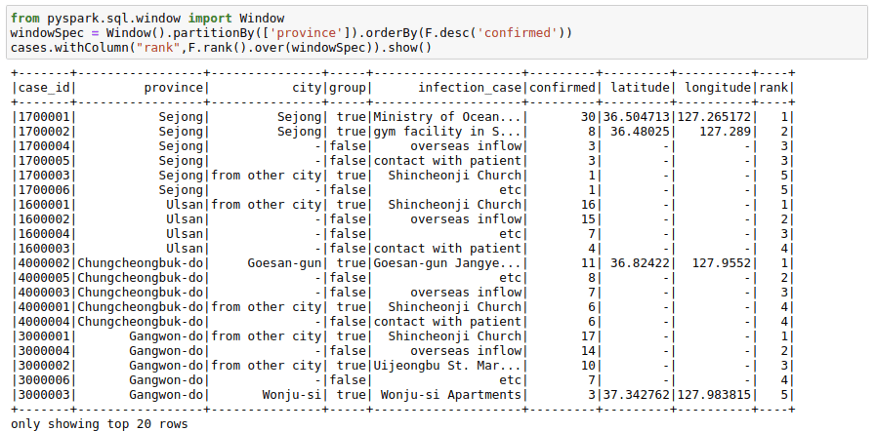
You can get rank as well as dense_rank on a group using this function. For example, you may want to have a column in your cases table that provides the rank of infection_case based on the number of infection_case in a province. We can do this by:
from pyspark.sql.window
import Window
windowSpec = Window().partitionBy(['province']).orderBy(F.desc('confirmed'))
cases.withColumn("rank",F.rank().over(windowSpec)).show()Lag Variables
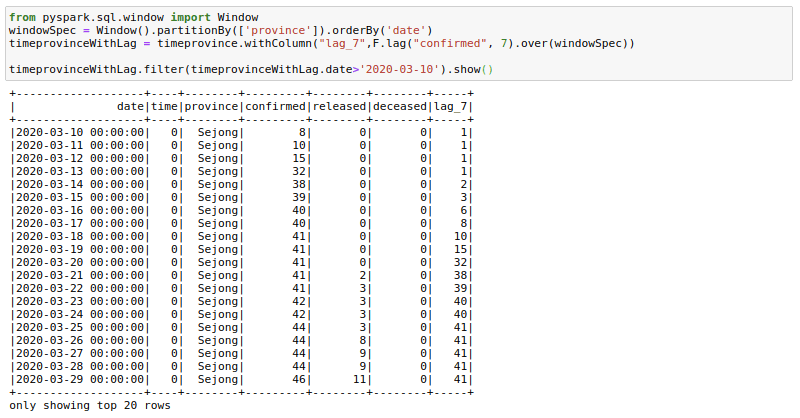
Sometimes our data science models may need lag-based features. For example, a model might have variables like the price last week or sales quantity the previous day. We can create such features using the lag function with window functions. Here I am trying to get the confirmed cases 7 days before. I am filtering to show the results as the first few days of corona cases were zeros. You can see here that the lag_7 day feature is shifted by 7 days.
from pyspark.sql.window import Window
windowSpec = Window().partitionBy(['province']).orderBy('date')
timeprovinceWithLag = timeprovince.withColumn("lag_7",F.lag("confirmed", 7).over(windowSpec))
timeprovinceWithLag.filter(timeprovinceWithLag.date>'2020-03-10').show()Rolling Aggregations
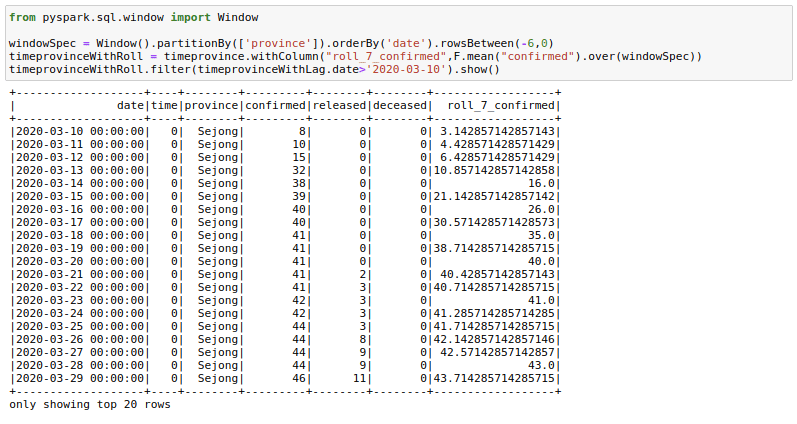
Sometimes it helps to provide rolling averages to our models. For example, we might want to have a rolling 7-day sales sum/mean as a feature for our sales regression model. Let us calculate the rolling mean of confirmed cases for the last 7 days here. This is what a lot of people are already doing with this dataset to see the real trends.
from pyspark.sql.window import Window
windowSpec = Window().partitionBy(['province']).orderBy('date').rowsBetween(-6,0)
timeprovinceWithRoll = timeprovince.withColumn("roll_7_confirmed",F.mean("confirmed").over(windowSpec))
timeprovinceWithRoll.filter(timeprovinceWithLag.date>'2020-03-10').show()There are a few things here to understand. First is the rowsBetween(-6,0) function that we are using here. This function has a form of rowsBetween(start,end) with both start and end inclusive. Using this we only look at the past 7 days in a particular window including the current_day. Here 0 specifies the current_row and -6 specifies the seventh row previous to current_row. Remember we count starting from 0.
So to get roll_7_confirmed for date 2020–03–22 we look at the confirmed cases for dates 2020–03–22 to 2020–03–16 and take their mean.
If we had used rowsBetween(-7,-1) we would just have looked at past 7 days of data and not the current_day.
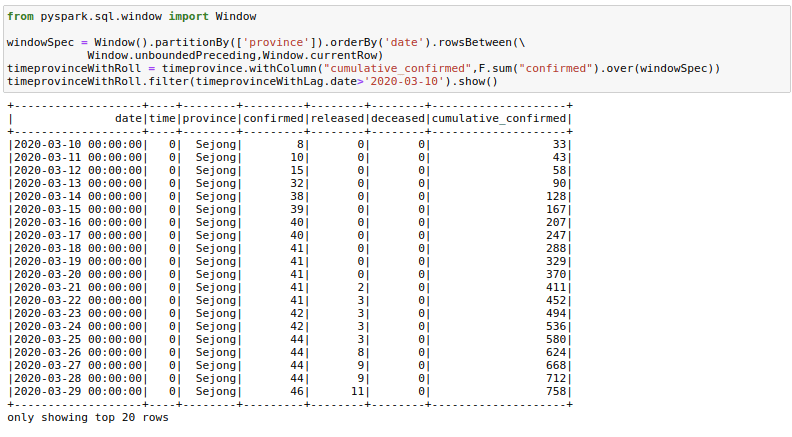
One could also find a use for rowsBetween(Window.unboundedPreceding, Window.currentRow) where we take the rows between the first row in a window and the current_row to get running totals. I am calculating cumulative_confirmed here.
from pyspark.sql.window import Window
windowSpec = Window().partitionBy(['province']).orderBy('date').rowsBetween(Window.unboundedPreceding,Window.currentRow)
timeprovinceWithRoll = timeprovince.withColumn("cumulative_confirmed",F.sum("confirmed").over(windowSpec))
timeprovinceWithRoll.filter(timeprovinceWithLag.date>'2020-03-10').show()6. Pivot Dataframes
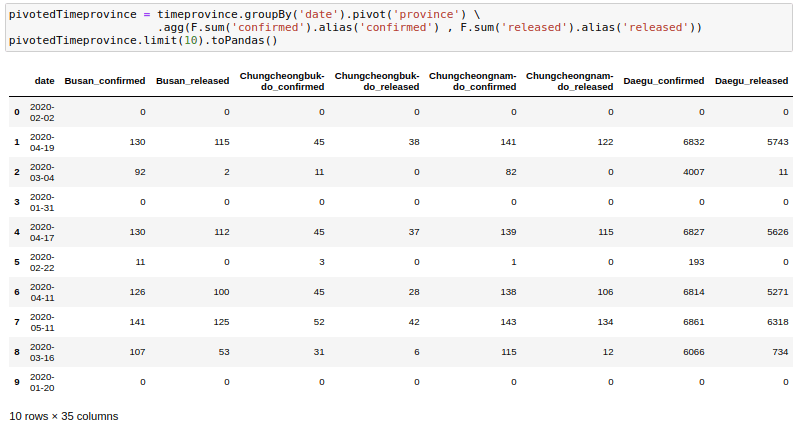
Sometimes we may need to have the dataframe in flat format. This happens frequently in movie data where we may want to show genres as columns instead of rows. We can use pivot to do this. Here I am trying to get one row for each date and getting the province names as columns.
pivotedTimeprovince = timeprovince.groupBy('date').pivot('province').agg(F.sum('confirmed').alias('confirmed') , F.sum('released').alias('released'))
pivotedTimeprovince.limit(10).toPandas()One thing to note here is that we need to provide an aggregation always with the pivot function even if the data has a single row for a date.
7. Unpivot/Stack Dataframes
This is just the opposite of the pivot. Given a pivoted data frame like above, can we go back to the original?
Yes, we can. But the way is not that straightforward. For one we will need to replace - with _ in the column names as it interferes with what we are about to do. We can simply rename the columns:
newColnames = [x.replace("-","_") for x in pivotedTimeprovince.columns]
pivotedTimeprovince = pivotedTimeprovince.toDF(*newColnames)Now we will need to create an expression that looks like the below:
.
.
.'Seoul_released' , Seoul_released,'Ulsan_confirmed' , Ulsan_confirmed,'Ulsan_released' , Ulsan_released) as (Type,Value)"The general format is as follows:
"stack(<cnt of columns you want to put in one column>, 'firstcolname', firstcolname , 'secondcolname' ,secondcolname ......) as (Type, Value)"It may seem daunting, but we can create such an expression using our programming skills.
expression = ""
cnt=0
for column in pivotedTimeprovince.columns:
if column!='date':
cnt +=1
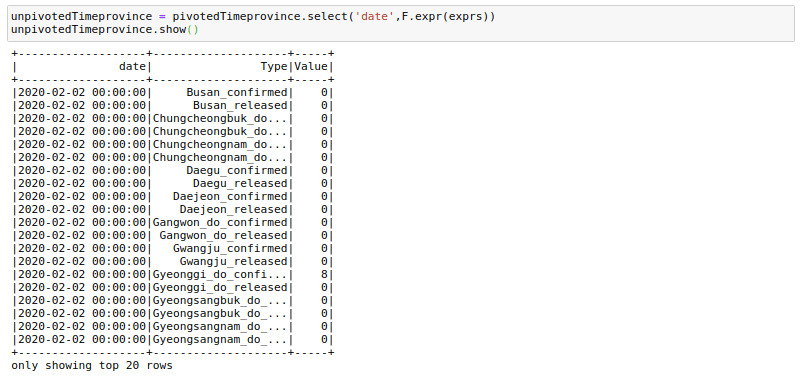
expression += f"'{column}' , {column},"expression = f"stack({cnt}, {expression[:-1]}) as (Type,Value)"And we can unpivot using:
unpivotedTimeprovince = pivotedTimeprovince.select('date',F.expr(exprs))And voila! we have got our data frame in a vertical format. There are quite a few column creations, filters, and join operations needed to get exactly the same format as before, but I will not get into those.
8. Salting
Sometimes it might happen that a lot of data goes to a single executor since the same key is assigned for a lot of rows in our data. Salting is another way that helps you to manage data skewness.
So assuming we want to do the sum operation when we have skewed keys. We can start by creating the Salted Key and then doing a double aggregation on that key as the sum of a sum still equals the sum. To understand this assume we need the sum of confirmed infection_cases on the cases table and assume that the key infection_cases is skewed. We can do the required operation in two steps.
1. Create a Salting Key
We first create a salting key using a concatenation of the infection_case column and a random_number between 0 to 9. In case your key is even more skewed, you can split it into even more than 10 parts.
cases = cases.withColumn("salt_key", F.concat(F.col("infection_case"), F.lit("_"), F.monotonically_increasing_id() % 10))This is how the table looks after the operation:
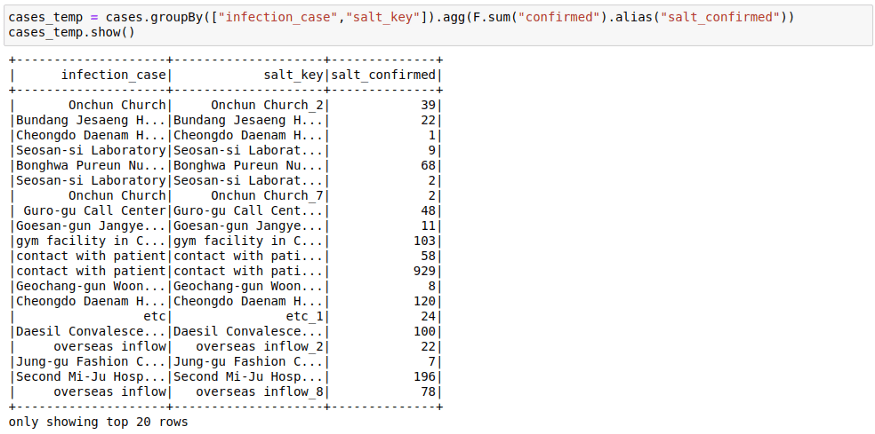
2. First Groupby on salt key
cases_temp = cases.groupBy(["infection_case","salt_key"]).agg(F.sum("confirmed")).show()3. Second Group On the original Key
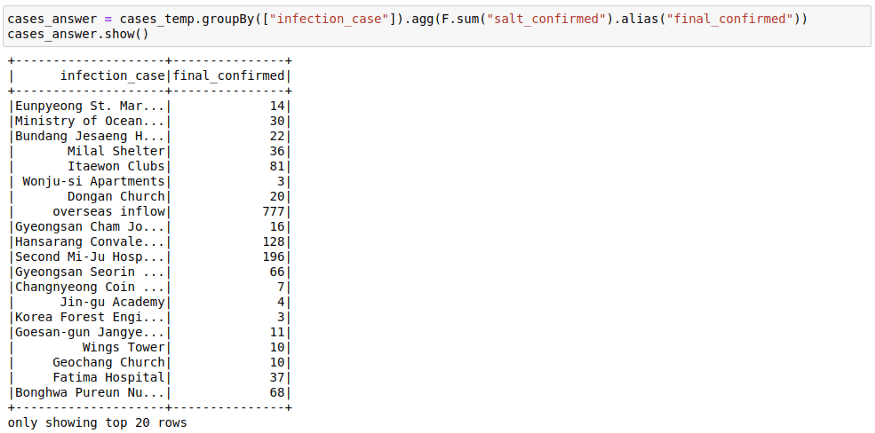
Here we saw how the sum of sum can be used to get the final sum. You can also make use of facts like:
- min of min is min
- max of max is max
- sum of count is count
You can think about ways in which salting as an idea could be applied to joins too.
Miscellaneous
Caching
Spark works on the lazy execution principle. What that means is that nothing really gets executed until you use an action function like the .count() one on a data frame. And if you do a .count function, it generally helps to cache at this step. So I have made it a point to cache() my data frames whenever I do an .count() operation.
df.cache().count()Save and Load from an Intermediate Step
df.write.parquet("data/df.parquet")
df.unpersist()
spark.read.load("data/df.parquet")When you work with Spark you will frequently run with memory and storage issues. While in some cases such issues might be resolved using techniques like broadcasting, salting, or cache, sometimes just interrupting the workflow and saving and reloading the whole data frame at a crucial step has helped me a lot. This helps spark to let go of a lot of memory that gets utilized for storing intermediate shuffle data and unused caches.
Repartitioning
You might want to repartition your data if you feel your data has been skewed while working with all the transformations and joins. The simplest way to do it is by using:
df = df.repartition(1000)Sometimes you might also want to repartition by a known scheme as this scheme might be used by a certain join or aggregation operation later on. You can use multiple columns to repartition using:
df = df.repartition('cola', 'colb','colc','cold')You can get the number of partitions in a data frame using:
df.rdd.getNumPartitions()You can also check out the distribution of records in a partition by using the glom function. This helps in understanding the skew in the data that happens while working with various transformations.
df.glom().map(len).collect()Reading Parquet File in Local
Sometimes you might want to read the parquet files in a system where Spark is not available. In such cases, I normally use the below code:
from glob import glob
def load_df_from_parquet(parquet_directory):
df = pd.DataFrame()
for file in glob(f"{parquet_directory}/*"):
df = pd.concat([df,pd.read_parquet(file)])
return dfTo deep dive into more details of PySpark, please check PySpark Tutorial section.